Methods

Multiplexed EV Immunoassays
Biotinylated capture antibodies are each coupled to one of ten unique U-PLEX® Linkers, which self-assemble onto unique array elements (or “spots”) on the U-PLEX plate. EVs presenting target surface antigens recognized by the capture antibodies bind to the associated spots. Bound EVs are detected using detection antibodies conjugated with electrochemiluminescent labels that recognize detection antigen(s) on the EVs. MSD’s electrochemiluminescence detection technology uses SULFO-TAG™ labels that emit light upon electrochemical stimulation initiated at the electrode surfaces of MULTI-ARRAY® microplates.
Tetraspanin Assays for Total EVs
U-PLEX arrays of capture antibodies targeting tetraspanin proteins CD9, CD63, CD81 and a negative control antibody are paired with a cocktail of detection antibodies for all three tetraspanin targets. Previously we showed these multiplexed arrays performed the same as the individual capture antibodies. Purified EVs from a known cell line are used as a calibrator to normalize signals between plates and experiments. The signals produced by these assays are proportional to the total EV concentration in the samples, and the control antibody is used to assess non-specific binding of EVs to the plate and ensure reliability of the data.
Cell Conditioned Media Samples
PC-3 (ATCC® CRL-1435™) and HCT-116 (ATCC® CCL-247™) cell lines were cultured at MSD using complete medium and at ATCC using EV-depleted medium. An hTERT-immortalized mesenchymal stem cell line (ATCC® SCRC-4000™) was cultured at ATCC in serum-free medium. Primary mesenchymal cells were cultured at Creative Biolabs in serum-free medium. Primary preadipocytes from subcutaneous adipose tissue and omental adipose tissue were grown at ZenBio in serum-free medium. EVs were measured in the raw conditioned media and in purified EV fractions prepared as described in the following section.
Sample Purification
Conditioned Medium from cell lines grown at ATCC were subjected to tangential flow filtration (TFF) at ATCC to purify and concentrate EVs. These materials are now available as products from ATCC: ATCC® CRL-1435-EXM™, ATCC® CCL-247-EXM™, and ATCC® SCRC-4000-EXM™. Conditioned medium from each of the sources were subjected to several purification methods at MSD: ultrafiltration by centrifugal filter units, precipitation by a volume exclusion reagent, size-exclusion chromatography using sepharose CL-2B. Primary preadipocyte CCM was also purified by ultracentrifugation at ZenBio.
Results

Comparing Total EV in each cell conditioned medium and EV isolate
Multiplex tetraspanin EV assays were used to assess the total EV content of each CCM sample and EV isolate. Signals from each EV isolate were divided by the signal in the most concentrated sample (HCT116 TFF) to produce a an relative EV concentration, compared to that sample. These estimated concentrations were only weakly dependent on which tetraspanin capture antibody was used. We used the average of the three markers to adjust data for overall EV concentration in the following tables. SEC produced much lower concentrations than the other methods as it diluted rather than concentrated the samples.
Assessing EV Phenotypes by Multiplex Surface Marker Profiling
All samples listed above were assayed for EVs with sixty-six surface markers including the three tetraspanins discussed above. The assays were divided into 10-plex panels, each panel including capture antibodies for 9 surface markers and a negative control antibody. A mixture of labeled antibodies for the three tetraspanins was used for detection. Signals for each marker were expressed as a percentage of the average of the CD9, CD63 and CD81 capture signals for that sample, approximating the percentage of the EVs in the sample expressing this marker. Values were omitted from the table where the specific capture produced less than 3 times the signal produced by the control antibody. EVs with thirty of the surface markers were detectable in at least one of the samples. Phenotypes were remarkably consistent across various samples from each cell line.
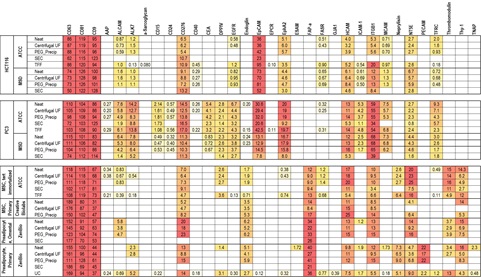
Unsupervised Clustering Analyses
Surface marker profile data from the preceding table were analyzed using two unsupervised clustering analyses. Hierarchical clustering, performed on log-transformed data using the complete linkage method, showed that EVs from the same cell line tended to show the same surface marker phenotypes independent of purification method or source. EVs from immortalized MSCs, primary MSCs and preadipocytes all had similar surface phenotypes and clustered together, except for the SEC method which produced low EV concentrations often near or below the assay detection limits. A second clustering approach, t-distributed stochastic neighbor embedding (t-SNE), carried out on non-transformed data was used to reduce the dimensionality of the data and allow the samples to be plotted in a 2-D space that captures the relationships between neighboring points. As seen with hierarchical clustering, the plots generated using t-SNE also showed the clustering of sample by cell line and type. The results demonstrated that our assay format can provide phenotypical information that is independent of EV purification method.

Conclusion
We developed multiplex EV surface marker assays and demonstrated their use for multimarker EV phenotyping. This flexible format enables rapid assay development for new EV subpopulations with or without sample purification. These results also demonstrate EV surface marker phenotyping via multiplex ECL assays may be used to distinguish EV populations from various cell types, and characterize bias introduced by purification.