2. Methods
MSD’s electrochemiluminescence detection technology uses SULFO-TAG labels that emit light upon electrochemical stimulation initiated at the electrode surfaces of MULTI-ARRAY and MULTI-SPOT microplates.
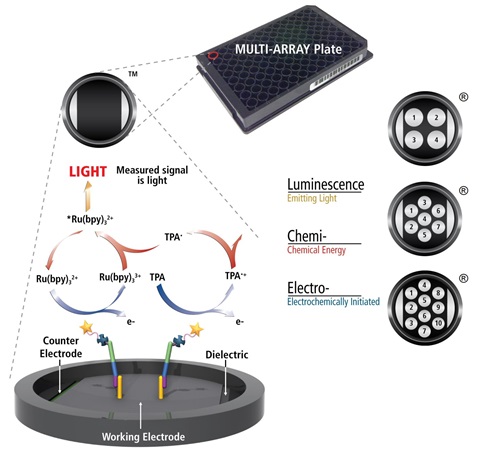
Electrochemiluminescence Technology
- Minimal non-specific background and strong responses to analyte yield high signal-to-background ratios.
- The stimulation mechanism (electricity) is decoupled from the response (light signal), minimizing matrix interference.
- Only labels bound near the electrode surface are excited, enabling non-washed assays.
- Labels are stable, non-radioactive, and directly conjugated to biological molecules.
- Emission at ~620 nm eliminates problems with color quenching.
- Multiple rounds of label excitation and emission enhance light levels and improve sensitivity.
- Carbon electrode surface has 10X greater binding capacity than polystyrene wells.
- Surface coatings can be customized.
Basis of the N-PLEX platform
N-PLEX plates contain 10 unique capture oligonucleotides that are bound to their corresponding spots on the electrode surface. Detection of a nucleic acid sequence is accomplished by hybridization of one or more probes with sequence complementary to these capture oligos and the nucleic acid of interest, followed by detection via electrochemiluminescence (i.e. biotin/streptavidin SULFO-TAG interactions). Blocking, hybridization, and detection are completed using proprietary MSD buffers and diluents.
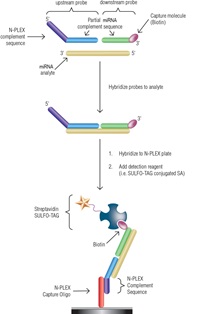
miRNA detection via two-probe approach
The two-probe detection assay used probes that were each complementary to the nucleotides of one-half of the miRNAs. The upstream probes contain spot-specific sequences at the 5′ ends that allow for hybridization to the N-PLEX plates. The use of spot-specific sequences enables detection of up to 10 distinct miRNAs per well. The downstream probes contain biotin on the 3′ ends for detection. The probes were hybridized to the miRNA and then to spot-specific capture oligos on the N-PLEX plates. SULFO-TAG labeled streptavidin was then used to detect the captured miRNAs.
Preparation of tissue lysate samples
Brain tissues were harvested from cynomolgus monkey (N = 1), mouse (N = 4), and rat (N = 2), and homogenized in lysis buffer. Lysates were collected and heat treated at 55°C for 30 minutes. No extraction step or amplification was necessary.
3. Results
Target miRNAs in Panel
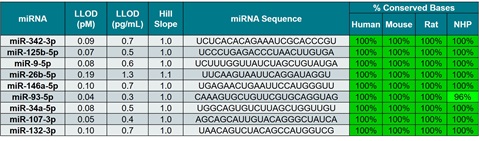
Figure 1. Probes were designed against several putative Alzheimer’s miRNA biomarkers and were run in a multiplex probe system. The selected miRNAs displayed high sequence similarity across species, with 96%–100% concordance among human, mouse, rat, and NHP. Calibration curves were run using synthetic miRNA templates, and the lower limit of detection (LLOD) was determined. The optimized assays were able to identify target miRNAs with a LLOD of lower than 1.27 pg/mL.
Sample Testing
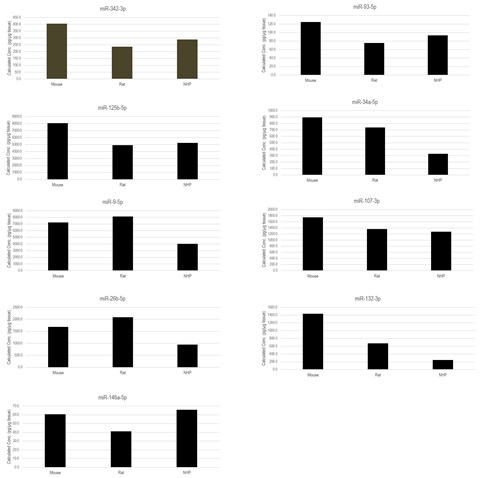
Figure 2. Lysed brain tissue samples from mouse, rat, and cynomolgus monkey were generated as follows: tissue samples were homogenized, heat-treated, spun down, and supernatants were collected. Twenty microliters of the lysed sample was used in the assay without further extraction. A multiplexed mixture of the upstream and downstream probes was hybridized to corresponding miRNAs in the samples. We showed that the miRNAs could be directly quantified from various tissue lysates without any complex extraction steps or amplification.
Specificity
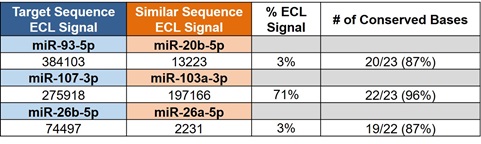
Figure 3. miRNAs that have similar sequences to the target miRNAs of interest were identified using BLAST. Each of the three similar miRNAs were produced synthetically and tested in the N-PLEX assays to evaluate specificity. ECL signals from the similar miRNAs were ≈ 3% of the target signal except for miR-103a-3p, which had an especially high percentage of conserved bases to miR-107-3p and displayed 71% of target signal (all miRNAs were tested at 125 pM).
Reproducibility

Figure 4A. Reproducibility of a multiplexed set of 9 targets over 5 plate runs were evaluated across 3 days. 96% of controls and 98% of LOQ samples were recovering within ±20% or ±25% of assigned control concentration, respectively (Figure 4B). All data points in brain lysate samples displayed high reproducibility, with 99%of data points falling within ±20% of the average calculated concentration (Figure 4B).
4. Conclusion
A multiplexed miRNA assay has been demonstrated that uses a simple, rapid workflow (<4 hours to result). It does not require RNA extraction and does not use a polymerase-based detection system. In a single 96-well plate run, the assay can detect nine miRNAs in 40 tissue lysate samples, providing a method that is both quantitative for multiple miRNAs and that has the potential for high-throughput screening.
Figure 2. Lysed brain tissue samples from mouse, rat, and cynomolgus monkey were generated as follows: tissue samples were homogenized, heat-treated, spun down, and supernatants were collected. Twenty microliters of the lysed sample was used in the assay without further extraction. A multiplexed mixture of the upstream and downstream probes was hybridized to corresponding miRNAs in the samples. We showed that the miRNAs could be directly quantified from various tissue lysates without any complex extraction steps or amplification.