Scientific Poster
>
Validation of Novel Multiplexed Serology Assays for Detection of IgG Antibodies against SARS COV 2 Key Variants
Validation of Novel Multiplexed Serology Assays for Detection of IgG Antibodies against SARS-COV-2 Key Variants
Download ↓
1. Introduction
COVID-19 is a viral illness caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). SARS-CoV-2 codes for various structural proteins, including the highly immunogenic Spike protein (S) and the Nucleocapsid protein (N) that plays a key role in virus transcription and assembly. Following the emergence of the virus, a wide range of serological tests were developed to estimate its seroprevalence, guide booster vaccinations, and select patients for anti-SARS-CoV-2 antibody therapies. Serological analysis is also an essential tool for understanding the immune correlates of protection against future COVID-19 waves following natural infection or vaccination. Despite a rapid increase in the number and availability of serology assays that can detect antibodies against SARS-CoV-2, there is limited knowledge about the performance and validation status of these assays. In addition, most are low throughput and only measure response to a single antigen, limiting their utility in capturing the breadth of antibody responses to SARS-CoV-2 and its variants.
Herein, we present validated, quantitative, multiplexed serology assays for the Nucleocapsid protein and key variants of SARS-COV-2 Spike and its receptor-binding domain (RBD) across two panels. The assays utilize 10-spot, 96-well plates coated with SARS-COV-2 antigens and an electrochemiluminescent (ECL) detection system to simultaneously detect IgG antibodies to SARS-CoV-2 Spike and RBD variants, including Alpha, Beta, Delta, and Omicron, in human serum. The panels were evaluated for accuracy, precision, analytical sensitivity, specificity, dilution linearity, and short-term stability. Clinical sensitivity and specificity were assessed using human serum from prepandemic samples. The validated assays accurately quantify IgG antibodies against RBD, Spike protein, Nucleocapsid protein and key variants of Spike protein (BA.2, BA.5, BA.2.12.1, BA.2.75, B.1.1.7, B.1.351, B.1.617.2; AY.4 and B.1.1.529; BA.1) and RBD (Omicron; BA.1, Omicron; BA.2, Alpha, Delta, Beta, BA.2.12.1, BA.2.75 and BA.4; BA.5). The multiplex SARS-CoV-2 ECL serology assays allow for sensitive, high throughput, and simultaneous measurement of IgG levels to multiple antigens in human sera, supporting its use in assessing exploratory endpoints for clinical trials.
 View More
View More
 View Less
View Less
2. MSD Platform
MSD Technology
MSD’s electrochemiluminescence detection technology uses SULFO-TAGTM labels that emit light upon electrochemical stimulation initiated at the electrode surfaces of MULTI-ARRAY® and MULTI-SPOT® microplates.
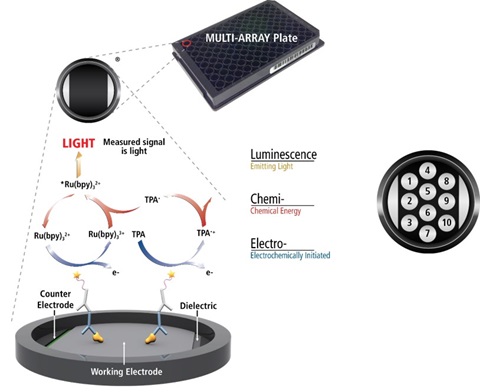
Electrochemiluminescence Technology
-
Minimal non-specific background and strong responses to analyte yield high signal-to-background ratios.
-
The stimulation mechanism (electricity) is decoupled from the response (light), thereby minimizing matrix interference.
-
Only labels bound near the electrode surface are excited, enabling non-washed assays.
-
Labels are stable, non-radioactive, and directly conjugated to biological molecules.
-
Emission at ~620 nm eliminates problems with color quenching.
-
Multiple rounds of label excitation and emission enhance light levels and improve sensitivity.
-
Carbon electrode surfaces have 10X greater binding capacity than polystyrene wells.
-
Surface coatings can be customized.
3. Method
The V-PLEX® COVID-19 Serology Kits quantitatively measure antibodies to antigens related to SARS-CoV-2 including variants of the SARS-CoV-2 virus, SARS, MERS, circulating Coronaviruses, and other respiratory pathogens. Plates are provided with antigens on spots in the wells of a 96-well plate. Antibodies in the sample bind to the antigens on the spots and anti-human antibodies (IgG, IgM, or IgA) conjugated with MSD SULFO-TAG are used for detection. The plate is read on an MSD® instrument, which measures the light emitted from the MSD® SULFO-TAG.
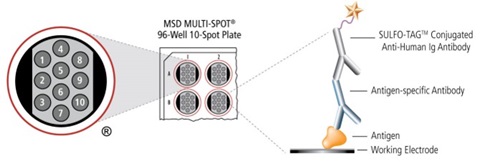
4. Results
a. Accuracy, Precision & Sensitivity
Inter-assay (between runs) and intra-assay (within runs) accuracy and precision were assessed by performing 6 runs on 3 different days by 2 different analysts. Each run consisted of standard curves, quality control (QC) samples, Upper Limit of Quantification (ULOQ), and Lower Limit of Quantification (LLOQ) samples. SARS-CoV-2 Key Variant Spike and RBD panels demonstrated good accuracy and precision. Inter-assay and intra-assay QC accuracy were ±25% and precision <20%, respectively (representative data for Mid QC is shown). The ULOQ and LLOQ of the assay are the highest and lowest known concentrations, at which precision was ≤25%, accuracy of 70%-130%, and total error of ≤40% are achieved within and between the established runs.
b. Assay Specificity
Specificity was evaluated by testing human serum samples collected pre- and post-COVID. Sixteen pre-COVID samples (collected in 2017) and sixteen post-COVID samples (collected in 2022) were tested at 5000-fold dilution.
IgG levels in most pre-COVID samples were BLQ. All post-COVID samples had precision <20% and quantifiable levels of IgG for all antigens. Higher levels of IgG were observed in post-COVID compared to pre-COVID samples indicating good specificity of the assays.
c. Parallelism & Prozone Effect
Parallelism and the presence of a prozone effect in human sera were assessed in 8 individual donor samples. Each sample was tested in serial dilutions to yield an 8-point titration (i.e., 100, 625, 1,250, 2,500, 5,000, 10,000, 20,000, and 40,000-fold dilution). The dilution-adjusted concentrations across the dilution series were assessed to evaluate SARS-CoV-2 IgG detectability and to characterize matrix interferences. Percent recovery was measured as the mean concentration at each dilution relative to the mean concentration at 5000-fold dilution. Good dilution linearity was observed between 625 to 40,000-fold dilutions in the Spike panel and 2,500 to 20,000-fold dilution in the RBD panel (%Recovery range ±30% is highlighted in green). The Hook effect was observed in a few serum samples at 100-fold dilution.
d. Selectivity
Selectivity was assessed in 10 individual human serum samples to evaluate matrix interference. In addition, 3 individual hemolyzed and lipemic human serum samples were tested to evaluate impact of hemolysis and lipemia on the assay, respectively. Each sample was diluted 5,000-fold and spiked with a reference standard. Spiked and unspiked samples were tested in duplicate. Spike recovery was calculated as measured concentration divided by nominal (endogenous and spike) concentration.
Matrix effect
No discernable matrix effect: >80% of samples recovered within ±25% across both panels.
Effect of Hemolysis and Lipemia
To evaluate the impact of lipemia and hemolysis on assay selectivity, three samples were tested after spiking with triglyceride-rich lipoprotein (1,500 mg/dL) and hemolysate (1,000 mg/dL). The samples were further spiked with a reference standard. Data indicates that most assays are tolerant to hemolysis and lipemia.
e. Short Term Stability
Experiments were performed to assess the stability of samples throughout the course of laboratory procedures. Human serum samples (n=3) were analyzed after being subjected to storage at RT (23°C ± 2°C) for 0, 2, 4, and 24 hours or 2-8°C for 0, 4, 24, and 48 hours. Freeze/Thaw Stability was assessed through 0, 1, 3, and 5 freeze-thaw cycles.
The data indicates that the samples are stable through storage at RT for 24 hours and at 2-8°C for 48 hours The samples were stable for up to 5 freeze-thaw cycles. Percent recovery was ±25% of baseline sample concentration and precision was ≤20%.
5. Conclusions
We validated quantitative, multiplexed serology assays that accurately and precisely quantify IgG antibodies against the antigens in SARS-CoV-2 Key Variant Spike Panel 1 (BA.2, BA.5, BA.2.12.1, BA.2.75, B.1.1.7, B.1.351, B.1.617.2; AY.4, B.1.1.529; BA.1, COV-2 S and COV-2 N) and SARS-CoV-2 Key Variant RBD Panel 1 (Omicron; BA.1, Omicron; BA.2, Alpha, Delta, Beta, BA.2.12.1, BA.2.75, BA.4; BA.5, COV-2 S1 RBD, and COV-2 N). The multiplex SARS-CoV-2 ECL serology assays allow for sensitive, high-throughput, and simultaneous measurement of IgG levels to multiple antigens in human sera, supporting its use in assessing exploratory endpoints for clinical trials. The assays were validated in MSD’s Bioanalytical Laboratory, which is GLP-compliant and provides preclinical and clinical sample testing services.
Customer Service/Orders
Scientific/Technical Support
Instrument Support
Company Headquarters