Scientific Poster
>
Use of the N-PLEX Platform for the Detection of Antisense Oligonucleotides (ASOs) in Plasma - ASGCT 2020
Use of the N-PLEX Platform for the Detection of Antisense Oligonucleotides (ASOs) in Plasma
Download ↓
1. Introduction
The number of FDA-approved drugs that are revolutionizing treatment strategies for cancer and other diseases is on the rise. One of the categories of drugs that is gaining prominence is antisense oligonucleotides (ASOs), which are modified, single-stranded DNA or RNA molecules (generally less than 30 nucleotides). These ASOs are complementary to specific mRNA sequences and alter production of the various corresponding proteins. Due to their short length, however, it is difficult to measure ASOs in circulation for PK studies. HPLC-MS/MS is frequently used to detect ASOs in biological samples, which provides good specificity but low sensitivity. Hybridization ELISA techniques have also been described and have better sensitivity, but greater sensitivity is still needed in many cases to enable thorough PK studies.
We sought to address this limitation by developing methodology around MSD’s newly developed nucleic acid detection platform, N-PLEX. The primary goal was to establish technical solutions that paired highly robust molecular techniques with detection on the N-PLEX platform to allow for highly sensitive detection of ASOs through chemiluminescence. One of the most common biological matrices that is used in PK studies, including ASOs, is plasma. Therefore, this methodology was developed and optimized for detection of ASOs in plasma samples, in which sub-pM detection was achieved in all three techniques discussed: OLA-mediated detection, T4 DNA ligase mediated detection, and detection via an RNase Protection Assay.
 View More
View More
 View Less
View Less
2. Methods
MSD® Technology:
MSD’s electrochemiluminescence detection technology uses SULFO-TAG labels that emit light upon electrochemical stimulation initiated at the electrode surfaces of MULTI-ARRAY and MULTI-SPOT microplates.
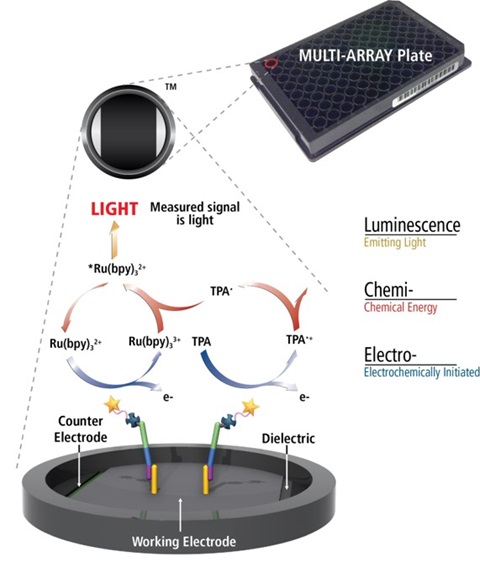
Electrochemiluminescence Technology
- Minimal non-specific background and strong responses to analyte yield high signal-to-background ratios.
- The stimulation mechanism (electricity) is decoupled from the response (light signal), minimizing matrix interference.
- Only labels bound near the electrode surface are excited, enabling non-washed assays.
- Labels are stable, non-radioactive, and directly conjugated to biological molecules.
- Emission at ~620 nm eliminates problems with color quenching.
- Multiple rounds of label excitation and emission enhance light levels and improve sensitivity.
- Carbon electrode surface has 10X greater binding capacity than polystyrene wells.
- Surface coatings can be customized.
Basis of the N-PLEX Platform
N-PLEX plates contain 10 unique capture oligonucleotides, each of which is bound to its corresponding spot on the electrode surface. Detection of a nucleic acid sequence of interest is accomplished by hybridization of one or more probes with sequence complementary to these capture oligos and the nucleic acid of interest, followed by detection via electrochemiluminescence (i.e., biotin/streptavidin interactions with SULFO-TAG). Blocking, hybridization, and detection are completed using MSD proprietary buffers and diluents.
Model ASO
For the development of methodology for ASO detection on the N-PLEX platform, a model DNA oligo was used for each of the techniques assessed. The model ASO that was chosen for this study is a 20-mer, DNA oligo with the following sequence: 5′ GGCTAAATCGCTCCACCAAG 3′.
Plasma Samples
Naïve mouse (CD-1 strain) plasma samples in sodium citrate were purchased from BioIVT (Westbury, NY). For all experiments involving the use of plasma, exogenous ASO was spiked into a high calibrator (Cal-1) and serially diluted in mouse plasma.
ASO Detection with Amplification Via the Oligonucleotide Ligation Assay (OLA)
The oligonucleotide ligation assay (OLA) used two probes that were complementary to the first or last 10 nucleotides of the ASO. One probe contained a spot-specific sequence at the 5′ end that allowed for hybridization to the N-PLEX plates, while the other probe contained a phosphate group on the 5′ end and a biotin on the 3′ end for ligation and detection, respectively. The thermostable Taq DNA ligase was used for multiple rounds of ligation of the two probes when appropriately hybridized to the ASO. The ligated probes were hybridized to spot-specific capture oligos on the N-PLEX plates to allow for detection via streptavidin bound to SULFO-TAG.
ASO Detection Via T4 Ligase-mediated Ligation
T4 ligase-mediated detection of the ASO used the same probes as OLA-mediated detection. However, T4 DNA ligase was used and only allowed for one round of ligation of the two probes when appropriately hybridized to the ASO. As with OLA, the ligated probes were hybridized to spot-specific capture oligos on the N-PLEX plates to allow for detection via streptavidin bound to SULFO-TAG.
ASO Detection Via RNase Protection Assay
The RNase protection assay utilized a single chimeric probe for the detection of the model ASO on the N-PLEX platform. This chimeric probe contained a 5′ DNA sequence that was complementary to the plate-bound capture oligo followed by an RNA portion that was complementary to the model ASO and a biotin on the 3′ end for detection via streptavidin bound to SULFO-TAG. Plasma samples were pretreated with RNAsecure reagent (ThermoFisher Scientific) and heated to 60 °C for 10 minutes to inactivate endogenous RNases. Once the probe was hybridized to both the ASO and the plate, RNase I was added to degrade any single stranded RNA sequence. Therefore, any RNA in the probe not fully protected by the ASO was degraded and the biotin released from the DNA portion of the probe, rendering it undetectable via streptavidin bound to SULFO-TAG.
3. Results: OLA-mediated ASO detection
Optimized Protocol
- A 10-point calibration curve was generated by spiking ASO into buffer or mouse plasma, serially diluting with 4-fold dilutions (166 pM top of curve), and hybridized with probes and Taq DNA ligase. Four blanks were used for estimated LLOD (LLODest).
- OLA was performed with the following protocol: 2 min at 95 °C and 30 cycles of 30 sec at 95 °C and 5 min at 37 °C. N-PLEX plates were blocked for 30 min at 37 °C during OLA cycling.
- Plate was washed, 50 µL of OLA product in hybridization buffer was added to the plate in duplicate, and plate was incubated for 1 hr. at 37 °C.
- Plate was washed, detection solution was added (50 µL per well), and plate was incubated for 30 min at room temperature.
- Plate was washed, read buffer was added (150 µL per well), and plate was analyzed with an MSD instrument.
Figure 1. A calibration curve was generated using the ASO, and detection was completed via OLA paired with N-PLEX methodology (see schematic and method description above). Results for calibration curves and LLODest are shown with the ASO in buffer or plasma.
4. Results: T4 Ligase-mediated ASO detection
Optimized Protocol
- A 10-point calibration curve was generated by spiking ASO into buffer or mouse plasma, serially diluting with 4-fold dilutions (166 pM top of curve), and hybridizing with probes and T4 DNA ligase. Four blanks were used for LLODest.
- Upstream and downstream probes were ligated together with T4 DNA Ligase (30 min at room temp). N-PLEX plates were blocked for 30 min at 37 °C during ligation.
- Plate was washed, 50 µL of T4 product in hybridization buffer was added to the plate in duplicate, and plate was incubated for 1 hr. at 37 °C.
- Plate was washed, detection solution was added (50 µL per well), and plate was incubated for 30 min at room temperature.
- Plate was washed, read buffer was added (150 µL per well), and plate was analyzed with MSD instrument.
Figure 2. A calibration curve was generated using the ASO, and detection was completed via T4 ligase paired with N-PLEX methodology (see schematic and method description above). Results for calibration curves and LLODest are shown with the ASO in buffer or plasma.
5. Results: RNase Protection Assay
Optimized Protocol
-
A 10-point calibration curve was generated by spiking ASO into buffer or mouse plasma, serially diluting with 4-fold dilutions (166 pM top of curve). RNAsecure pretreatment inactivated plasma RNases.
-
ASO was hybridized with the chimeric probe. Four blanks were used for LLODest. N-PLEX plates were incubated for 30 min at 37 °C during hybridization.
-
Plate was washed, 50 µL of product in hybridization buffer was added to the plate in duplicate, and plate was incubated for 1 hr. at 37 °C.
-
Plate was washed, RNase I was added to degrade RNA portion of probe if not hybridized to the ASO, and plate was incubated for 30 min at 37 °C.
-
Plate was washed, detection solution was added (50 µL per well), and plate was incubated for 30 min at room temperature.
-
Plate was washed, read buffer was added (150 µL per well), and plate was analyzed with MSD instrument.
Figure 3. A calibration curve was generated using the ASO, and detection was completed via RNase protection assay paired with N-PLEX methodology (see schematic and method description above). Results for calibration curves and LLODest are shown with the ASO in buffer or plasma.
6. Results: Reproducibility Testing
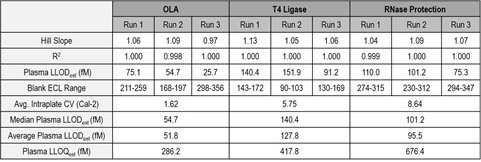
Table 1. Using the methodology described in figures 1-3, assays were run in mouse plasma for 3 consecutive days for each method to assess the reproducibility of the data with the model ASO. Highly reproducible results were obtained for each method described.
7. ASO Detection Methodology Comparisons
* Assay time could potentially be shortened with further optimization for specific ASO
These data indicate that, paired with various techniques, the N-PLEX platform is a viable option for highly sensitive detection of ASOs in complex biological matrices. In fact, an average plasma LLODest of 51.8, 127.8, and 95.5 fM was achieved using OLA, T4 DNA ligation, and RNase protection assay, respectively. Factors to consider when designing assays for the detection of ASOs on the N-PLEX platform include the length and modifications of the specific ASO, type of biological matrix, assay time, and requirements for sensitivity and specificity.
Customer Service/Orders
Scientific/Technical Support
Instrument Support
Company Headquarters